2021
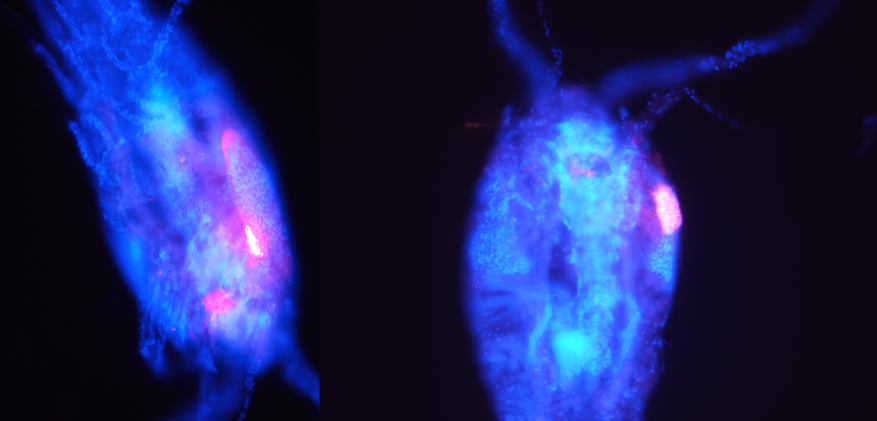
Male differentiation in the marine copepod Oithona nana reveals the development of a new nervous ganglion linked to Lin12-Notch-Repeat protein-associated proteolysis .
Sugier K, Laso-Jadart R, Kerbache S, Kafer J, Arif M, Bertrand L, Labadie K, Martins N, Orvain C, Petit E, Poulain J, Vacherie B, Wincker P,
Jamet JL, Alberti A, Madoui MA. Biology. 2021; 10(7):657. https://doi.org/10.3390/biology10070657
Estimating the travel time and the most likely path from Lagrangian drifters.
O'Malley M, Sykulski AM, Laso-Jadart R, Madoui MA. Journal of Atmospheric and Oceanic Technology, 2022; 38(5), 1059-1073, doi: https://doi.org/10.1175/JTECH-D-20-0134.1
The first AI-based mobile application for antibiotic resistance testing.
Pascucci M, Royer G, Adamek J, Aristizabal D, Blanche L, Bezzarga A, Boissinot D,
Boniface-Chang G, Brunner A, Curel C, Dulac-Arnold G, Malou N, Nordon C, Runge V,
Samson F, Sebastian E, Soukieh D, Vert JP, Ambroise C, Madoui MA. 2021. Nature Communications 12, 1173 (2021). https://doi.org/10.1038/s41467-021-21187-3.
2020
metaVaR: introducing metavariant species models for reference-free metagenomic-based population genomics.
Laso-Jadart R, Ambroise C, Peterlongo P, Madoui MA. Plos One. 2020. e0244637. https://doi.org/10.1371/journal.pone.0244637
Investigating Population-scale Allele Specific Expression in Wild Populations of Oithona similis (Cyclopoida, Claus 1866).
Laso-Jadart R, Sugier K, Petit E, Labadie K, Peterlongo P, Ambroise C, Wincker P, Jamet JL, Madoui MA. Ecology and Evolution. 2020; 10: 8894– 8905
2019
Discovering millions of plankton genomic markers from the Atlantic Ocean and the Mediterranean Sea.
Arif M, Gauthier J, Sugier K, Iudicone D, Jaillon O, Wincker P, Peterlongo P,
Madoui MA. Molecular Ecology Resources. 2019 Mar;19(2):526-535
Reference genome for pea provides insight into legume genome evolution. Kreplak J, Madoui MA, Cápal P, Novák P, Labadie K, Aubert G, Bayer PE, Gali KK, Syme RA, Main D, Klein A, Bérard A, Vrbová I, Fournier C, d'Agata L, Belser C, Berrabah W, Toegelová H, Milec Z, Vrána J, Lee H, Kougbeadjo A, Térézol M, Huneau C, Turo CJ, Mohellibi N, Neumann P, Falque M, Gallardo K, McGee R, Tar'an B, Bendahmane A, Aury JM, Batley J, Le Paslier MC, Ellis N, Warkentin TD, Coyne CJ, Salse J, Edwards D, Lichtenzveig J, Macas J, Doležel J, Wincker P, Burstin J. A Nat Genet. 2019 Sep;51(9):1411-1422. doi: 10.1038/s41588-019-0480-1. Epub 2019 Sep 2. PMID: 31477930.